Summary
- Capa, Deviations, Quality Events
- CAPA and Deviations
- New QMS? Pay attention to these keys success factors!
- Validation of the Imaging Technology for a novel microbiological colony counter
- Barrier Technologies & Revision of Annex 1: requirements & operational responses for manufacturers
- GMP Annex 1 and Lyophilization
Validation of the Imaging Technology for a novel microbiological colony counter
Most microbiologists would claim that the recorded number of colony forming units (CFU) were data. However, that number is someone’s (skilled technician) interpretation of the number of colonies on the plate. Experience has shown that different technicians (each skilled) can and frequently do observe different counts on the same sample and the data recorded in the lab notebook or batch record. However, these are the best data available to us.
Validation Imaging Technology microbiological colony counter
It must also be remembered that the CFU is only an estimate of the number of cells present. It is a skewed estimate at best, as the only cells able to form colonies are those that can grow under the conditions of the test (e.g., incubation media, temperature, time, oxygen conditions).
Even those do not represent a single cell, but rather those that happened to be well separated on the plate and so can be distinguished after growth. A colony could arise from one cell or several thousand cells, Sutton(1).
The ability to accurately count colonies depends on many factors e.g., acuity of the analyst vision, lighting conditions, size and color contrast of the colony against the media color. In a multi-center study by Paris et al(2) a very good analysis was performed looking at the critical parameters using beads of different sizes and colors distributed around petri plates.
From the 3 criteria influencing the detection accuracy, the most significant appears to be:
1. A clear correlation between the beads size and the detection performance. Below 212-250 μm up to 74% of incorrect results are measured, and the smaller the beads, the worse the enumeration error level is.
2. Colony location was also a factor, 212-250 μm beads located at the middle or the edge of the plate demonstrated a significant reduction of accurate counts, from 91.20% to 67.20%.
3. The influence of contrast was less significant but for beads around 212-250 μm there was better detection with higher contrast.
Based on those results, an accurate limit of detection for the human eye is close to 250 μm. Under this threshold, the percentage of false negatives increases significantly.
The use of membrane filtration to recover organisms has lower working ranges due to smaller surface area than with a 90mm Petri plate. ASTM provides countable ranges of 20-80 CFU/membrane, 20-200 CFU for spread plates, and 30-300 CFU for pour plates (3). The US Food and Drug Administration Bacterial Analytical Manual (BAM) recommends 25-250 CFU/plate as a countable range (4).
Within the ranges quoted the ability of the operator to accurately count the colonies deteriorates as numbers get higher. The inability to detect merged colonies that occur more frequently on a crowded plate also affects accuracy. A guidance to compare the ability of analysts to count colonies is given in APHA Standard Methods for the Examination of Dairy Products 85, the analyst counts must not differ 90% of the time by more than 10% from the mean count performed by 3 independent analysts.
The goal of the paper is to validate that an automated colony counter can address the key require- ments to accurately enumerate colonies on a growth media while minimizing the negative effects of some of the variables listed above.
1. Colony counter technology
To improve accuracy and repeatability for colony enumeration, recent advances in Pharmaceutical microbiology have seen the introduction of technologies designed to automate the enumeration of colonies on the surface of media plates or other support matrices. The illumination and detection systems used in these technologies vary from full spectrum white light bulbs through narrower wavelength LEDs to very narrow wavelength lasers. Data capture is usually by a Photo Multiplier Tube (PMT) or a Charge Coupled Detector (CCD) with some form of wavelength selecting filter(s) through a lens, that may magnify the image. The technology used will in each case detect microbes as either single cells, microcolonies or full colonies and through the use of software algorithms separate true microbial counts from non-microbial objects that could cause false positives. The accuracy of the technology depends on the ability of the software to interpret the images of a wide range of microbial colony shapes, colors and sizes with a range of background materials that may confuse the detection algorithms. The growth characteristics of organisms can also create problems due to cells clumping or growing together or swarming rapidly over the plate. With the importance for the accurate enumeration of any microbial contaminant in a pharmaceutical product it is critical that the accuracy of the enumeration be determined during the validation process.
The technology used for the detection of microbial microcolonies uses a black mixed ester cellulose membrane to support growth and reduce background fluorescence during colonial growth on traditional microbiology media substrates.
The detection of events on the membrane surface is a combination of physical size and fluorescent intensity. The fluorescent intensity is primarily driven by the cell volume (that contains the flavin fluorophore). Robust yeast cells with large volumes can be detected in smaller colonies, ~10 cells, than the slim rod bacteria, ~100 cells.
The Growth Direct technology uses the natural autofluorescence of microorganisms to facilitate their detection on the membrane surface. The details of the technology are described by London et al (6).
2. Vision validation strategy
Accuracy of the result is the key parameter to assess the performance of the vision system and associated software. However, to assess the accuracy a suitable control method needs to be defined. From the end user viewpoint, the “gold Standard” is the cfu result that a trained and experienced analyst would obtain given a cassette with growing colonies. There are some drawbacks to this approach:
1. The ability of analysts to count colonies can be variable through training level and visual acuity
colonies can merge and by performing the traditional endpoint read at day 5 to 7 the analyst could call the combined entities a single colony. 2. Colonies increase in size with time so a count on day 3, 5 or 7 may differ significantly due to the degree of overgrowth seen.
3. if the colony is very small at the end point it may be lost against the media background, especially if the colony is semi transparent, and be invisible to all analysts.
Using multiple analysts to read each test plate will improve the accuracy and be a good assessment of the analysts “control” count.
A more definitive assessment of the accuracy of the system can be made by utilizing the Growth Direct® software to extract each image in the test series and visually check the decisions made by the software for each of the developing colonies at each timepoint. This would be defined as the “Reference Method”.
3. Parameters Potentially Affecting the Growth Direct® Advanced Imaging System
In order to validate that the technology accurately enumerates microorganisms, examples of the full spectrum of variables that may be seen need to be tested. The following factors are among those that were evaluated:
1. Media type may affect detection, TSA (with EM derivatives), SDA and R2A
2. Cellular Volume: Representative size range for bacteria, yeast and mold. Extremes for bacterial cells would be B. diminuta and B. megaterium. The Growth Direct® system cannot detect a single cell but uses micro colonies, the detection of which. The detection of a micro colony depends on the number of cells and the volume of the cells (large volume increases flavin content) to have sufficient fluorescent signal for detection. 3. Colony morphology: e.g., shape, margins, elevation, size, texture, pigmentation and general optical properties
4. Color: transparent, colorless, cream, tan, white, yellow, orange and red
5. Colony size: pinpoint, small, medium and large
6. Pigment types: different organisms contain a range of pigment types that may affect detection
7. Application type: the source of the sample can affect the colony types detected.
To validate the accuracy of enumeration, test cassettes were run for all the test species defined in the tables below. Sufficient cells were used to generate a colony number that can be easily counted by eye, 1 to 150 cfu. The colonies on a media cassette were enumerated by the Growth Direct system and on the same cassette by multiple analysts, usually 3, immediately after incubation on the system.
4. Enumeration accuracy
Accuracy was performed using the Environmental monitoring format:
EM format- organisms spread plated to the EM cassette membrane surface that is supplied in contact with the media, TSA LP80. Media cassettes were incubated on the system for 3 to 5 days at 20-25°C or 30-35°C.
At the end of the incubation the colonies on each of the test cassettes were enumerated by 3 independent operators. The mean counts for the operators were compared to the result generated by the system.
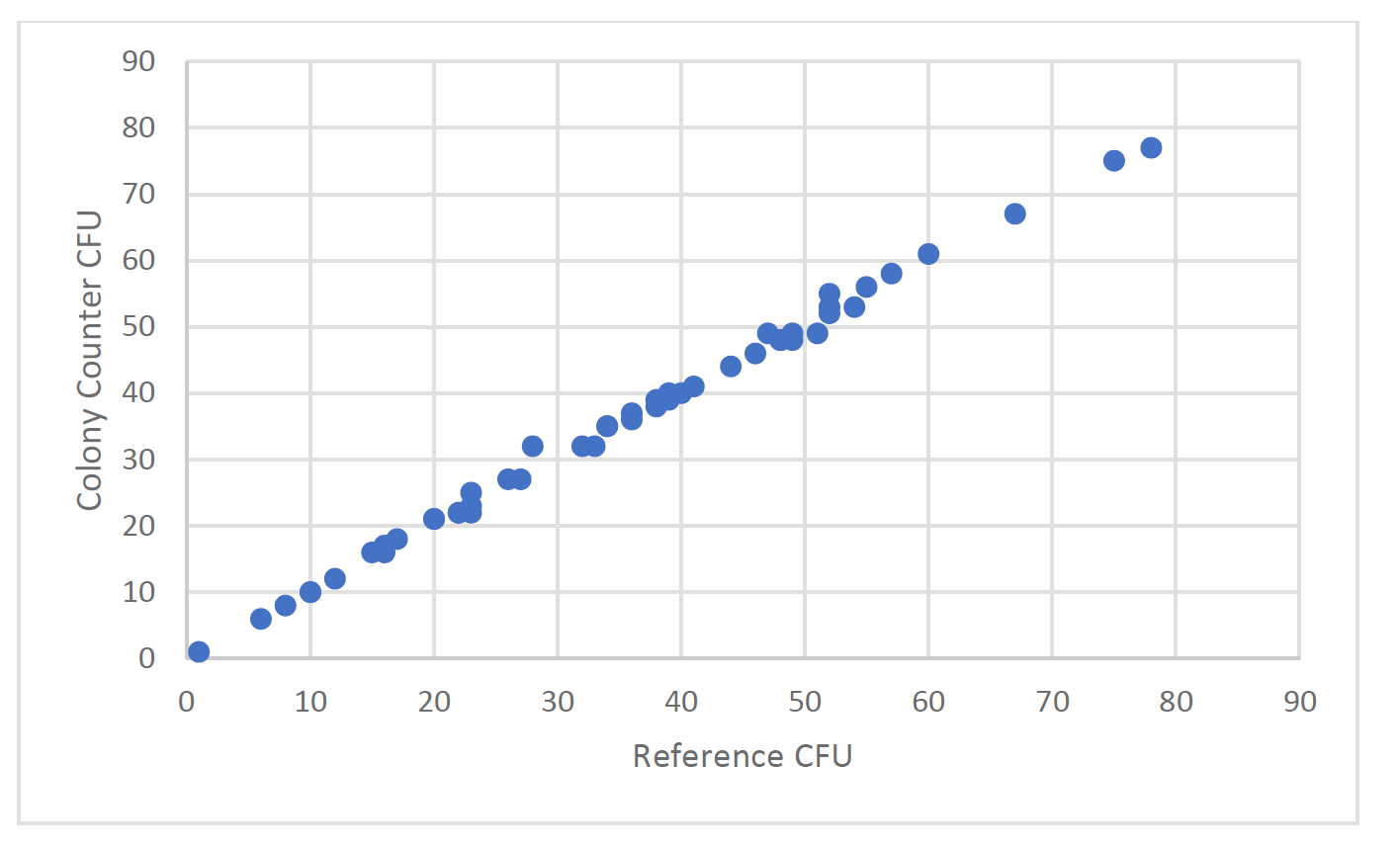
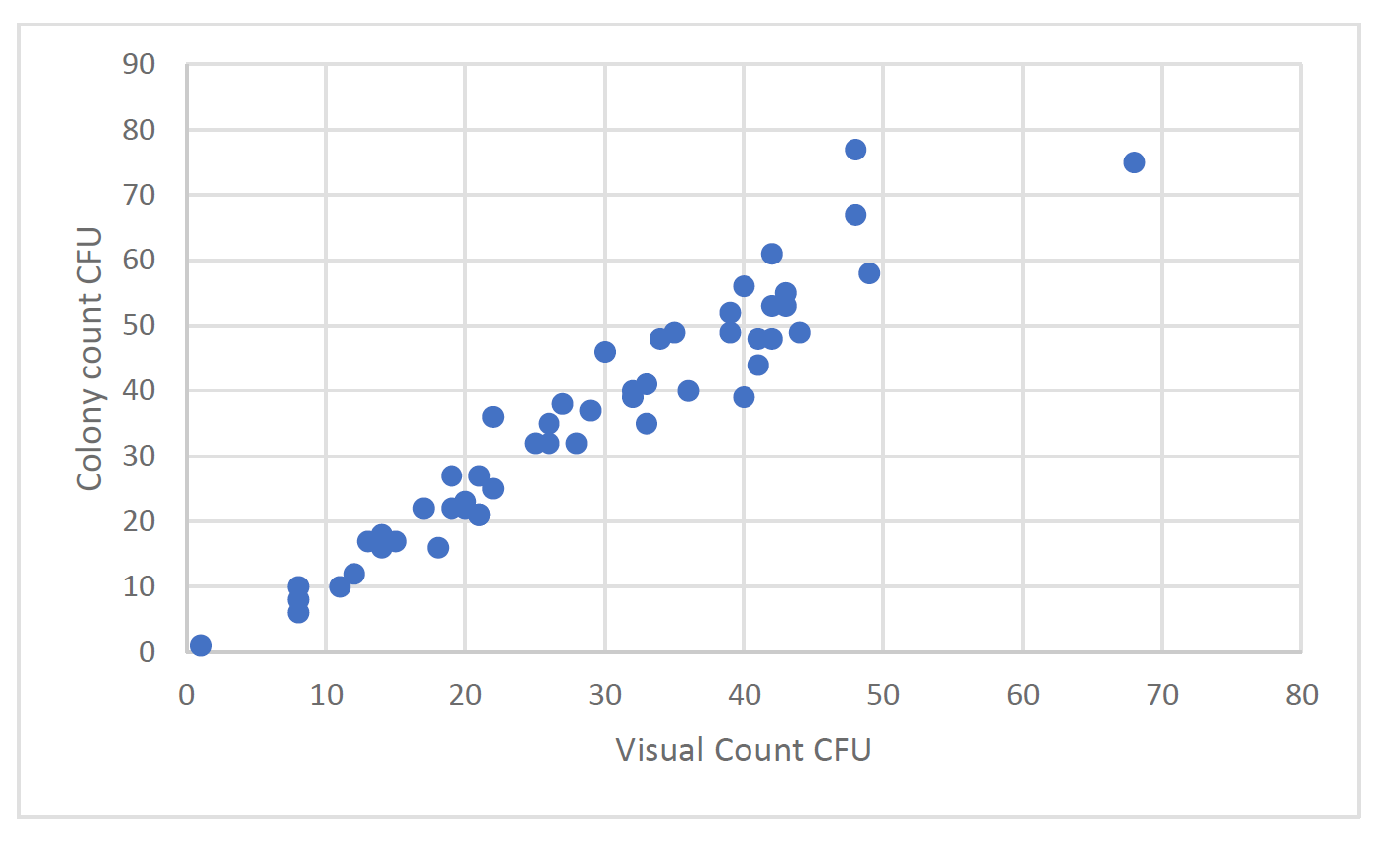
Overall, there is an excellent agreement between the reference method count and the counts obtained using the colony counter technology. As the technology is based on traditional microbiology there are still cases where colonies are located so close together that they merge before the algorithms can detect separate entities. Those mergers however are less frequent than would occur using the discriminatory power of the human eye. Figures 8a and 8b show the effect on accuracy with the introduction of the human eye for colony enumeration.
Figure 2a, Reference CFU compared to colony counter CFU from active air samples taken from an unclassified area adjacent to a clean room. Enumeration performed after Day 3 of incubation.
Figure 2b, mean visual CFU from 3 analysts compared to colony counter CFU. Samples from active air samples taken from an unclassified area adjacent to a clean room. Enumeration performed after Day 3 of incubation.
As can be seen from Figure 2 a and b, the accuracy of the human count is inferior to the automated colony counter. It should be noted that in the range of organisms more characteristic of a production facility, <<50cfu, the colony counter has extremely good accuracy compared with the reference count. At higher counts some inaccuracy can occur due to merger colonies.
5. Discussion
Colony counter technology has been an evolving science since the early models from the 1970’s. Improving illumination modules with more camera pixel optics gave better image discrimination that improved the detection of colonies. The early “end point” methods (read once at the end of incubation) were limited in the enumeration due to the growth characteristics that could merge colonies. A lower colony count than the true value would then be the result. The merged colonies were very difficult to isolate with software algorithms available at the time. Three improvements in this area have made a significant improvement to colony counter performance that facilitates its application to routine QC testing in the pharmaceutical industry:
• Use of natural autofluorescence by the colony allowed specific colony detection compared to visible light for colonies of the same color as the media or that occurred at the edge of Petri Plates.
• The use of machine learning/AI has enabled far better discrimination of the colonies from background noise or other objects.
• Use of a “kinetic” approach where images are taken at intervals through the incubation phase. This approach allows the discrimination of colonies before they merge thus giving a more accurate count.
The application of the technology to routine use has been encouraged by the addition of automation. Colony counting and the manual effort required to incubate, count and record the data has always been an issue for microbiology staff. More recent technologies have implemented the automated incubation with the colony count technology and associated secure data transfer to central databases or electronic batch records. Prior to use in a pharmaceutical environment the analytical method needs to be fully validated for its intended use. This document can be used as supporting information for the validation of the Growth Direct® system as the unit goes through the traditional Installation/Operational, Performance and Method Qualification (IOPMQ) process. The testing described in this paper was the result of a risk analysis performed to define what the key factors were that could adversely affect the performance of the test method. As a result, the data show that the system is a robust and accurate analytical tool for the enumeration of microbial colonies for the major applications used in the microbiology QC laboratory. The technology has been implemented globally for routine testing and has been successfully included in a number of regulatory drug applications.
Références :
- Sutton S, “How Many”, PMF Newsletter. 12 (9), 2006.
- Paris, A.; Plourde-Awobi, L.; From variable operator numeration to the standardized 3P station automated colony counting on environmental monitoring culture media plates. La Vague. 2020, 66, July, 27-34.
- ASTM, D5465-93 (1998) Standard Practice fur Determining Microbial Colony Counts from Water Analyzed by Plating Methods, 1998.
- FDA, Maturin, LJ and JT Peeler, “Chapter 3: Aerobic Plate Count, Bacteriological Analysis Manual, http://www.fda.gov/food/ Sd.enceResearchfLaboratoryMethodsfBacteriologicaiAnalyt icaiManualBAMfucrn063346.htm, (accessed June 28, 2011
- 2004 APHA Standard Methods for the Examination of Dairy Products
- London, R., J. Schwedock, A. Sage, H. Valley, J. Meadows, M. Waddington and D. Straus 2010. An automated system for rapid non-destructive enumeration of growing microbes. PLos ONE 5(1): e8609
Share
Validation Imaging Technology microbiological colony counter

David L. Jones