Summary
- Control strategy and robotics: the future of production sites?
- Robotized loading of cartridges at Lilly
- Small, flexible filling and packaging systems - using robots could mean great benefits
- Automated and instantaneous enumeration of viable microorganisms with Red One™, solid-phase cytometry platform
- Method analatycal performance strategy in commercial quality control laboratories
- “Replicate strategy*”: but what is it exactly?
- A3P International Congress 2020: Feedback and key messages from the Annex1 revision process panel discussion
- Regulatory compliance: a re-evaluation of strategic subcontracting relationships in the “new normal”
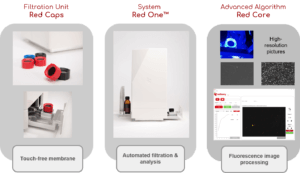
Automated and instantaneous enumeration of viable microorganisms with Red One™, solid-phase cytometry platform
In order to effectively control the quality of a pharmaceutical product, it is necessary to monitor and analyze microbiological contamination throughout its manufacturing process. The enumeration of viable microorganisms on agar culture media remains the reference method and a requirement by regulatory authorities for product release. There are many alternative methods that allow for faster results.
Currently however, they are based on using an incubation phase prior to testing on the rapid method platform, existing systems either use the early detection of micro-colonies (by fluorescence, auto- fluorescence or ATP bioluminescence) or an indirect measurement of metabolites resulting from microorganisms growing in the liquid phase (measurement of pH, CO2 or quantity of ATP).
Some technologies, such as flow cytometry or ATP assay after cell lysis, make it possible to quantify the microbial load or bioburden without a culture phase. However, these two methods do not reach the sensitivity required to detect the low microbial loads required to meet the specifications for pharmaceutical products or for in-process control (1 to 100 CFU typically).
Based on reduced incubation times, new alternative and rapid methods are currently in use in control laboratories. On the production side, “real-time” analyzers are now needed to monitor bioburden and manage contamination control more proactively.
The new generation of solid-phase cytometry technology developed by Redberry allows the detection of microbial load of 10 to 10E+5 CFU in volumes of 100 μL to 100 mL in 10 minutes. Sanofi Pasteur and Redberry have collaborated on the evaluation of a pre-commercial version of this technology, which will be available in 2021 for pharmaceutical applications.
Red OneTM, the automated solid phase cytometry technology
The Red OneTM detection platform is based on solid-phase cytometry technology [1], i.e. it is capable of detecting unicellular microorganisms captured and fixed on a polyester membrane (porosity 0,3 μm) after filtration of the sample. These microorganisms are then labeled with a cell viability staining agent (a derivative of fluorescein diacetate) which reacts to the presence of esterases, enzymes present in any type of microorganism having a metabolic activity.
This type of staining has been widely proven by some alternative methods in the past, whether for the detection of single cells [1] or micro-colonies [2]. It is in particular recognized for exhibiting minimal cellular toxicity. However, its implementation in current platforms requires an incubation phase for cell labeling (in order to guarantee the presence of sufficient material to be detected), an often-complex manual handling of the sample (resulting in extended time to result and increased risk of contamination).
The fluorescence emitted by the labeled cells is detected, with Red OneTM, leveraging on the latest generation optical technologies: high-power LED lighting for excitation (λexc = 485 nm) and very high- resolution CMOS sensor camera for emission (λem = 520 nm). These technologies make it possible to significantly reduce the size and cost of the equipment compared to systems put on the market twenty years ago, thus making it possible to use it as close as possible to a production line or in the laboratory and in complete biological safety (because it can be positioned under a laminar flow hood). All the operations necessary for the analysis (filtration and labeling) are fully automated and do not require the intervention of an operator.
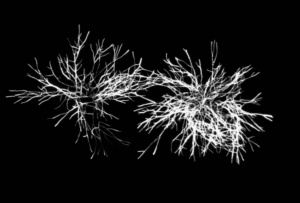
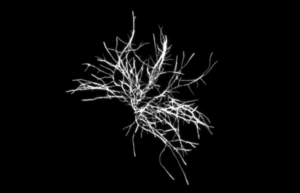
Finally, high-resolution images of the microorganisms are taken before, during and after the injection of the viability staining agent. Thus, a change in the fluorescence (or labeling kinetics) of each cell is recorded over time (total data recording time: 10 minutes). This kinetics is representative of the gradual assimilation of the viability staining agent by the microbial cell and corresponds to the diffusion time through the cell membrane and then to the enzymatic reaction. Analysis of this kinetics makes it possible to differentiate viable cells from inert particles (auto-fluorescent, for example) with a high level of reliability (Figure 2).
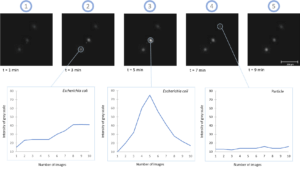
Figure 2.b: Detection of Aspergillus brasiliensis mycelium (NCPF 2275)
Figure 2: High Resolution fluorescence imaging and labeling kinetics
This approach makes the system less sensitive to background noise (see below), which avoids additional sample preparation steps, such as successive washes of the membrane or the use of counter-markers.
Objective of the study: evaluation of the sensitivity to background noise and detection performance (recovery rate)
The sensitivity of rapid methods to background noise from test matrices is an important performance criterion. Indeed, these methods often rely on the use of biochemical markers that, according to the principle of detection, are sensitive to the presence of nucleic acids, proteins, ATP or even enzymes. This can result in false positives which affect the usefulness of the system for routine applications.
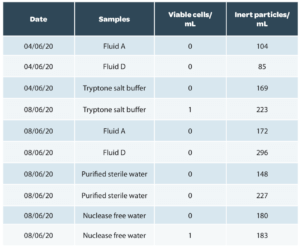
An example of results of analyses of various buffers and sterile rinsing solutions used in pharmaceutical microbiological control with Red OneTM is given below (Table 1.a) and on production matrices from Sanofi Pasteur (Table 1.b). These results confirm the ability of Red OneTM to correctly classify inert particles in most cases. They were obtained on pre-production consumables (caps) assembled in an aseptic environment but not sterilized by radiation, which explains the residual number of false positives detected at the time of testing.
The impact of these false positives has been evaluated by Redberry on a set of 203 water samples, buffers and sterile rinsing solutions: the microbiological load of the samples cannot be determined below 8.44 cells (mean plus 3 times the standard deviation of the counts) which is below the detection limit of the system.
Table 1: Examples of results on sterile samples and production matrices
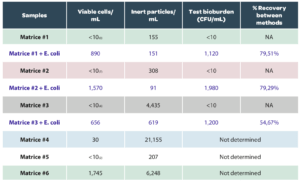
The performance of Red OneTM has been evaluated on several species of microorganisms (Table 2): using reference strains from the ATCC pharmacopoeia (BIOBALL® Multishot 550 – bioMérieux) and 4 environmental isolates.

- Counting with Red One TM: successive dilutions in tryptone salt to reach target concentrations between 10E+5 and 10 CFU/mL. Volume tested with Red OneTM: 1 mL.
- Inoculum control: culture method on TSA agar (bioMérieux), incubation for 2 days at 30-35°C.
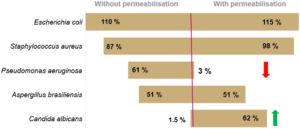
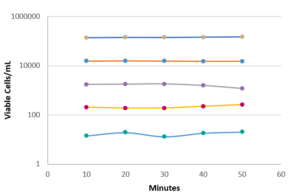
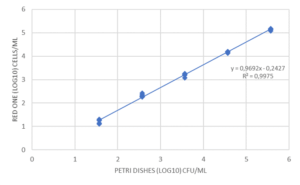
The recovery rate corresponds to the ratio of the count given by Red OneTM to that obtained on TSA agar. The recovery rates between the counts given by Red OneTM and those obtained on culture dishes are shown in (Figure 3):
- a satisfactory recovery rate (> 60%) for bacteria
- lower recovery rates for the yeast/fungal species Candida albicans and Aspergillus brasiliensis
The reason for reduced recovery of the yeast and fungal species may be associated with the marker having difficulty entering the cells.
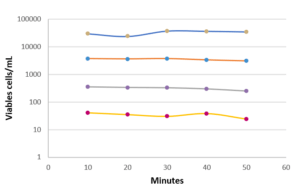
To improve the labeling, a membrane permeabilization solution has been developed by Redberry and tested by Sanofi Pasteur. It is applied by the operator just prior to analysis of the sample (1 mL of the permeabilization buffer is added to the filtered sample for 30 seconds). This step produces a very significant increase in the recovery rate for Candida albicans (1.5% to 62%), but it also has an adverse impact on those microorganisms with a more fragile membrane (e.g. Pseudomonas aeruginosa see Figure 3).
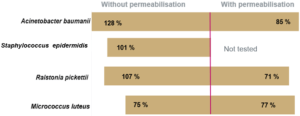
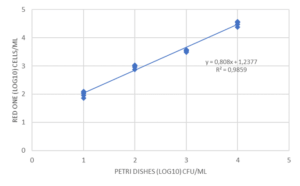
Finally, some tests were carried out to evaluate the repeatability and linearity of the method on Micrococcus luteus, Ralstonia picketti and Mycobacterium bovis. The results obtained on Mycobacterium bovis and Micrococcus luteus are detailed in Figure 4. The repeatability was assessed on samples prepared in serial dilutions (factor 10) with concentrations between 50 and 5.10E+4 CFU/mL. Each concentration was tested 5 times with an interval of 10 minutes between 2 measurements, the required time for an analysis.
The linearity over this measurement range is very high (R2> 0.98), as is the repeatability of the results obtained for each dilution tested. (Figure 4). This performance is due in particular to the automation of the detection method: no intervention of the operator is required for all operations up to the result of the analysis. Thus, the filtration conditions of the sample, the volume of marker injected, and the exposure time of the cells to labeling are precisely controlled by the equipment, with no delay between each step.
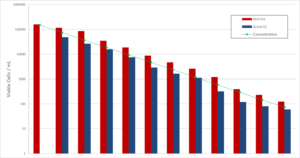
As shown in the example of Mycobacterium bovis, the counts given by Red OneTM were also compared to those given by flow cytometry (Accuri C6 – Becton Dickinson, Figure 6). Although the viability marker is different (here ChemChrome V6 – bioMérieux) with a preparation and incubation period at 30-35° C of 20 minutes, the results are similar up to a range of 10+2 to 10+4 CFU/mL, below which conventional flow cytometers are limited in terms of accuracy.
Conclusions and Future Potential
Solid-phase cytometry is a technology able to reach the low limits of detection required for pharmaceutical microbiological applications [1]. However, its use with the first generation of equipment developed at the beginning of the 2000s was proven to be complex, in particular because of the different sample preparation operations: successive washings, counter- labeling, reactivation phase, various manipulations of the membrane, and microscopic confirmation of the results for the detected events.
By implementing the staining kinetics in a fully automated system, Red OneTM overcomes all or part of these steps to get a reliable result in 10 minutes, rather than in a few hours or a few days, with very good repeatability. This allows potential use in “real-time” applications for which obtaining an instantaneous result is advantageous (piloting industrial processes, restarting production units or after cleaning checks for example). Counting bacteria of interest (management of banks for example) is also a possible application for which Red OneTM is suitable by giving an accurate and rapid count.
In order to meet the strict demands of the pharmaceutical industry, the future prospects of Red OneTM applications for quality control are as follows:
- the implementation of reactivation phase of the cells (3 hours) allowing the detection of microorganisms that are difficult to label (weak enzymatic activity or slow membrane permeabilization)
- the addition of an enrichment phase on the membrane (48 hours incubation step) to ensure the multiplication of microorganisms and reach a detection limit of 1 CFU (targeted application: release testing).
Acknowledgments – We warmly thank Khouloud Badri (Engineer from PolyTech Clermont-Ferrand) and Joseph Pierquin for their scientific contribution and for the writing of this article. We also thank Stéphanie Baron, David Pacory and Julien Arnaud for their technical support, as well as Jonathan Macron for his contribution to the success of this collaboration.
Partager l’article

Lucile PLOURDE-OWOBI – Sanofi Pasteur
Lucile is R&D Scientific Initiatives Manager and Lab Automation Network Leader at Sanofi Pasteur, missioned on “microbiome” initiatives to develop the Research and Industry partnerships and to accelerate scientific innovation. With 20 years of experience in industrial microbiology (analytical, manufacturing, research and development), Lucile is a SME in microbial and molecular methods, bio-banking, strain characterization and up-stream process. Developing alternative, rapid and automated technologies to improve lab productivity is a part of her journey, as well as the troubleshooting solving for industrial operations.
lucile.plourde@sanofi.com

Marine SCHNETTERLE – Redberry
Currently in charge of microbiological assays as part of the development of Red One™, a new generation of solid-phase cytometry platform designed by Redberry. Marine is more specifically responsible for the development of release testing workflows as well as reagent kits for the pharmaceutical quality control. Her role also involves support to roll-out and customers. Prior to joining Redberry, Marine obtained her PhD in bacteriology with the French Armed Forces Biomedical Research Institute (IRBA) in 2018. Her main work focus was on the role of efflux pumps in the antibiotic resistance of bacteria such as Burkholderia pseudomallei.
marine.schnetterle@redberry.net
Glossary
ATP: Adenosine Tri-Phosphate
LED: Light-Emitting Diode
CMOS: Complementary Metal Oxide Semiconductor
CFU: Colony-Forming Unit
References
[1] Comparative assessment of epifluorescence microscopy, flow cytometry and solid-phase cytometry used in the enumeration of specific bacteria in water, Karine Lemarchand, Nathalie Parthuisot, Philippe Catala, Philippe Lebaron, Aquatic Microbial Ecology, Vol. 25, pp 301–309, Septembre 28th 2001
[2] Fluorescence-Based Rapid Detection of Microbiological Contaminants in Water Samples, Hervé Meder, Anne Baumstummler, Renaud Chollet, Sophie Barrier, Monika Kukuczka, Frédéric Olivieri, Esther Welterlin, Vincent Beguin, and Sébastien Ribault, The ScientificWorld Journal, Volume 2012, Article ID 234858, 10 pages