Summary
- There is everything to gain from working in campaign mode, a breakthrough in isolator productivity
- Visual inspection: main findings of ANSM inspections
- Cleaning validation of production equipment: Visual inspection, accreditation of staff in “visually clean”
- A Refresher on Disinfectant Wet Contact Time
- THE EU MDR and IVDR : Combination products now subject to the same degree of surveillance as standalone medical devices
- Suitability of the Mono-Mac-6 cell line for the detection of endotoxins and non-endotoxin pyrogens
- Biological indicators, random growth. Is your decontamination cycle really at fault?
- Determining a Strategy for Container Closure Integrity Testing of Sterile Injectable Products
- Efficient Control Strategy enabled by structured Knowledge
- High-performance solution for real-time colony counts on filtration membranes in microbiological analysis with ScanStation
High-performance solution for real-time colony counts on filtration membranes in microbiological analysis with ScanStation
The enumeration of viable microorganisms on agar culture media remains the recommended reference method or imposed by the various standards for microbiological tests in release control. It involves either direct inoculation of the liquid sample to be analyzed (spreading on the surface of the medium or pour plate method), or the release of the solid sample biomass into a liquid volume which is subsequently filtered out onto a membrane (pore diameter 0.22 to 0.45 µm) which will then be deposited onto the appropriate culture medium.
Although proven, culture-based techniques can have different biases from sample preparation to final Petri dish (Corry et al., 2007). Looking more specifically at the drawbacks associated with final enumeration, we identify the main ones to be as follow:
- The time factor: counting is a long process where one or more technicians has to count each CFU and do this for a very important number of Petri dishes. This activity represents several hours per week in a microbiological analysis laboratory,
- The technician’s ability to detect colonies: this can be impacted by various single or combined causes. The phenotype of the colonies can make detection on a medium or membrane (translucent colonies or colonies with a color close to the membrane or medium). Colony size is also a constraint when there are micro colonies or in the early stages of development. Their location on the edge of Petri dishes can also lead to omissions of some colonies. Finally, a high density of colonies on the medium will also complicate the detection of each individual CFU.
- The phenomenon of confluence during incubation: As the plates are read at the end of incubation (from 24 hours to several days), some colonies may be masked by others or overlap, making their detection impossible and/or giving the impression of the presence of a single colony when in reality there are several colonies (false negative).
- The risk of counting errors: intrinsically linked to the two points mentioned above, the phenomenon of counting errors, particularly when the number of plates to be counted is large, is also recurrent and has been highlighted in numerous studies for several decades. (Fowler et al., 1978 ; Peeler et al., 1982).
- In addition to these risks of errors in counting at the bench, there is also the risk of errors related to the processing of the result, either involuntary or with the aim of masking non-compliance:
– Error in editing the result,
– Forgetting to edit the result,
– Falsification of data,
– Erasure of data,
– Data destruction.
In recent years, regulations have been striving to impose the notion of data integrity.
Data integrity refers to maintaining and ensuring the accuracy and consistency of data throughout its life cycle, from the design, implementation and use of any system that stores, processes or retrieves data. Data integrity is a key regulatory concern and to assist those working in the pharmaceutical and healthcare industries, documents have been published by the Food and Drug Administration (FDA, 2018). It has now become a fundamental requirement of the pharmaceutical quality system described in Chapter 1 of GMP. The data refer to the ALCOA system and must therefore be:
– Attributable: author and source identified,
– Legible: interpretable and long term,
– Contemporary: recorded at the time they are collected,
– Original or certified copy,
– Accurate
In this context and to overcome all these problems, in a logical evolution of semi-automatic or automatic detection systems that have appeared for a little over 20 years, the emergence of automated counting technologies with computerized management of data has emerged. Among these technologies, we will study the example of the ScanStation developed by Interscience.
1. ScanStation Overview
ScanStation, a real-time colony counting and incubation station, detects and counts colonies as soon as they appear for 100, 200 or 300 Petri dishes. ScanStation allows real-time microbial reading during incubation. It provides complete and anticipated microbial results for quality control and R&D laboratories in the pharmaceutical industry, integrating full data traceability in compliance with 21 CFR part 11.
ScanStation was designed and developed by Interscience, a French company created in 1979. The major innovation of ScanStation is to perform automatic colony counting at an early stage of their development, from the beginning of the incubation cycle. Petri dishes are photographed at regular intervals, every 30 minutes (every 60 minutes for the ScanStation 300 model), delivering a time lapse of the microbial growth. The final result is thus known up to three times faster than with a traditional colony count, eliminating repetitive work. The very first image taken (T0) also CFUs from artefacts present on the plate thus offering a more accurate and much earlier result. The accelerated growth video allows the operator to “go back in time” and helps to validate the results, as the operator can review the first images to determine the initial number of colonies, identified by a green cross.
2. Aim of the study : verification of the detection /count performance of colonies on filtration membranes by ScanStation
Although the detection of colonies on agar media has been validated, the question of detection performance on filtration membranes remains to be verified. First tests had been carried out by Interscience but only on cellulose membranes. Preliminary work within the framework of the Aptar pharma / Interscience collaboration has enabled software improvements to be made to detect colonies on nitrate, acetate and cellulose ester membranes commonly used in microbiological analyses. The objective of the following study is to verify the performance of the device for the detection of CFU on membranes, in particular by defining:
- The type of membrane and the counting parameters of the software allowing the best counting,
- Its ability to enumerate bacterial strains of different phenotypes,
- Its maximum detection threshold to ensure that it is sufficient to quantitatively count a large number of colonies close to 150 CFUs (value accepted as the maximum threshold that can be counted on membranes).
3. Materials & Methods
3.1 Tested membranes
Four references of membranes commonly used in microbiological controls are tested:
- White cellulose nitrate with grid, 0.45µm (Sartorius Microsart® part no.: 16D01–25-06–BK)
- White cellulose acetate membrane without grid, 0.45µm (Sartorius Order No.: 11106–47——–N)
- Black cellulose nitrate membrane with grid, 0.45µm (Sartorius part no.: 16D03–25-H6–BK)
- Black cellulose ester membrane without grid, 0.45µm (Millipore reference: HABP04700)
3.2 Microbial strains used
Seven strains are used, five of which are listed in the Pharmacopoeia:
- Aspergillus brasiliensis (mould) in BioBall 550 (bioMérieux reference: 56011)
- Bacillus subtilis (bacterium) in Bioball 550 (bioMérieux reference: 56012)
- Candida albicans (yeast) in BioBall 550 (bioMérieux reference: 56013)
- Pseudomonas aeruginosa (bacterium) in BioBall 550 (bioMérieux reference: 56017)
- Staphylococcus aureus (bacterium) in BioBall 550 (bioMérieux reference: 56019)
Two strains isolated from production environments (non-sterile areas) called U1 (translucent beige colonies) and U2 (white colonies).
The various tests consist of:
- A pure inoculum for each strain
- An inoculum containing S.aureus (33%), B.subtilis (33%) and P.aeruginosa (33%) (Consortium 1)
- An inoculum containing S.aureus (25%), B.subtilis (25%) and P.aeruginosa (25%) A.brasiliensis (25%) (Consortium 2)
3.3 Generation of inoculum and plating of membranes
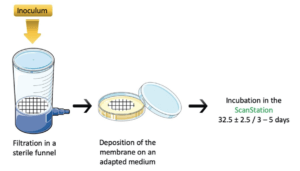
The membrane plating protocol is summarised in Figure 1
The primary inoculum is prepared and brought to the desired concentration. It is diluted in a volume of 100 mL of saline solution (0.9%), then poured into the filter funnel. This volume is filtered through the membrane. The membrane is then removed and deposited onto the appropriate culture medium: Tryptic Soy Agar (TSA) for bacteria and microbial consortia, and Sabouraud Dextrose Agar (SAB-agar) for yeast and molds strains in pure cultures. The media are incubated in the ScanStation at 32.5°C ±2.5 for 3 to 5 days.
3.4 Determination the « membrane / Scanstation detection program »
ScanStation has different incubation settings. These vary the lighting of the plates during photographic acquisition as well as the colony detection algorithm by the image processing software. There are three predefined membrane settings:
– White paper membrane
– White polycarbonate membrane
– Black membrane
An initial test carried out with S. aureus enables the ScanStation program to be adapted to the most optimal detection for each type of membrane.
A common inoculum is used and fractions of it are diluted in a volume of 0.9% sterile saline (100 mL). They are filtered through each type of membrane. These are then deposited on TSA-agar media and incubated in the ScanStation at 32.5±2.5°C for 3 days. Each test consists of a series of 5 membranes for each membrane reference.
A manual count is carried out in duplicate in order to determine the total number of colonies developed on each membrane using the images taken by the apparatus.
3.5 Determining the best combination of « membrane / ScanStation detection parameters »
For each ScanStation/membrane parameter pairs, detection tests are carried out with S. aureus, B. subtilis and the two factory strains U1 and U2. Inoculum and incubation are carried out as described in the previous paragraph. The series are also composed of 5 membranes.
A manual count is carried out in duplicate in order to determine the total number of colonies developed on each membrane using the images taken by the apparatus.
3.6 Maximum detection limit
Series of pure inoculum, of S.aureus, B.subtilis or P.aeruginosa, obtained by cascade dilution to 1/10th, were used to inoculate black membranes with grid (Sartorius). These are deposited on TSA-type agar culture medium and then incubated in the ScanStation as described above.
4. Results et discussions
The results obtained are converted into average detection percentages. Ideally to consider an effective detection the manufacturer sets a detection percentage of 95 ± 5%.
4.1 Determining the « Membranes / ScanStation detection program»
An initial series of tests is carried out to determine which program allows for the most accurate detection according to the membrane used. It should be noted that the “white polycarbonate membrane” parameter allows the most efficient colony detection when using white grid or membranes without grid (Table I) with detection percentages of 95 ± 3% and 98 ± 2% respectively. For black membranes, the “black membrane” parameters allows total detection. Conversely, the “white paper membrane” parameter does not allow detection of colonies despite the marked contrast between the color of the membrane (black) and that of the colonies (yellow) (Figure 2).

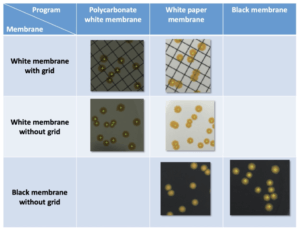
For the following tests the parameter “white polycarbonate filtration membrane” is used for detection on white membranes and the program “black filtration membrane” is used for detection on black membranes.
4.2 Choosing the most optimal « ScanStation/ membrane parameter» pair for colony detection
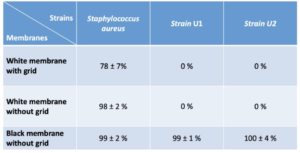
Three bacterial strains were tested in order to find out which parameter/membrane pair was the most efficient to detect colonies of very different phenotypes (Table II). For S.aureus, detections of the order of 99 ± 2 % and 98 ± 2 % were noted for the black and white membranes without grid respectively. For the white membranes with grid, the percentage of detection is lower than the other two with 78± 7 %. It should be noted that tests carried out with B.subtilis on white membranes, whether with grid or not, showed the same trend. Indeed, the detection rate goes from 70 ± 2 % on non-grid membranes to 19 ± 8 % on membranes with grid. Analysis of the images showed that the undetected colonies were systematically located on the grids. There is therefore a degrading effect of the grids on the detection performance, which must be taken into account when choosing white membranes.
For the two strains isolated from the factories, only the black membranes allow detection and colony count of the order of 99 ± 1 % for the E. coli type strain and 100 ± 4 %. No detection of these light-colored strains is observed on white membranes.
Considering these results, the choice of black membranes coupled with the ScanStation’s “black filtration membrane” parameter is the combination that seems to be the most suitable for detecting different phenotypes in a high-performance way.
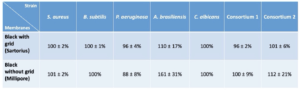
These results have been confirmed by other series of tests on two references of black filtration membranes (membranes without grid, Millipore and membranes with grid, Sartorius) with pure strains and consortium mimicking a mixed population as can be found on the cultures from the controls (Table III). The detection percentages on Sartorius and Millipore black membranes are comparable and respective of :
- 100 ± 2 % and 101 ± 2 % for S.aureus
- 100 ± 1 % and 100 % for B.subtilis
- 96 ± 4 % and 88 ± 8 % for P.aeruginosa
- 100 % for C.albicans
The detection percentages of P.aeruginosa are lower because the colonies are sometimes translucent. This phenotype can cause little contrast with the membrane, which can limit the software’s ability to detect certain colonies. Moreover, the analysis of the images showed that some of the uncounted colonies were close to another. This creates confluence and prevents the two colonies from being counted separately.
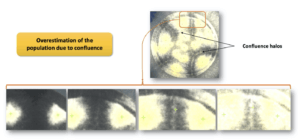
The percentages of detection of A. brasiliensis show a high variability of detection on both types of membranes, equal to 110 ± 17% (Sartorius membrane) and 161 ± 31% (Millipore membrane). This mold is white and invasive. Its ability to spread on the membrane creates a confluence between colonies. This increases the density of mycelium in the confluence areas and thus the white color is intensified. This color variation is detected by the software and a colony is then counted in error (Figure 3). The first photograph in Figure 3 shows a black membrane covered by colonies of A. brasiliensis. Confluence halos are visible.
For the counts of consortium 1 and 2, the detection percentages are:
- 96 ± 2% on Sartorius membranes and 100 ± 9% on Millipore membranes for Consortium 1
- 101 ± 6% on Sartorius membranes and 112 ± 21% on Millipore membranes for Consortium 2.
The counts of microbial consortium show similar percentages of detection. The difference between the actual number of colonies and the number of colonies counted is less than 5%. However, if the software analyzes the detections on the images, not all colonies are counted. This is a numerical compensation. A double counting error of one colony compensates for a single counting error of two nearby colonies. This compensation is frequent, especially when counting B. subtilis in the consortium. The error is minor but repeated during the analyses. Note that this type of error is quickly corrected by the final check of the technician on the screen at the end of incubation.
In the same way, the same phenomenon is observed for the detection of A.brasiliensis. The number of colonies seems to be more overestimated on Millipore membranes than on Sartorius membranes. It is difficult to explain this observation. One hypothesis is that the membrane may affect the phenotype of A.brasiliensis colonies. This difference in appearance could influence the detection of colonies by the ScanStation software.
4.3 Maximum detection/counting limit on membrane
The detection limit is the maximum number of colonies that can be quantitatively and reliably counted on a membrane by the ScanStation.
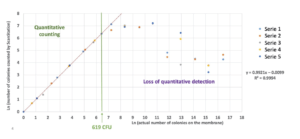
Manual counting is performed by completing the ScanStation count. The uncounted colonies are added to the value obtained automatically and multiple counts of the same colony are subtracted. The graphical compilation of the 5 independent series allows to define an area where a linearity is observed between the detection of colonies by the ScanStation and the actual number of colonies on the membrane (Figure 4). An extrapolation of the area of loss of linearity allows to define approximately a maximum detection limit of 619 colonies. Considering that beyond 150 colonies on a membrane, the membrane is considered to be non-accountable, this result shows a significant performance of the device well beyond the maximum values that can be used on filtration membranes.
To confirm the counting performance of the ScanStation when the number of colonies approaches the threshold value of 150 CFU, additional tests were carried out with inoculum of S.aureus, B.subtilis and P.aeruginosa. The concentration of these inoculums is adjusted to 150 CFU. The average enumeration percentages obtained are as follows:
- S. aureus : 100%
- B. subtilis : 98 ± 1%
- P. aeruginosa : 99 ± 2%
This confirms the capacity of the device to detect a large number of colonies of different phenotypes at maximum exploitable values.
4.4 Observed software errors
Different types of counting errors were made during our tests and thus identified to help Interscience improve its software interface. These observations are presented in table IV. The double colony count (161 occurrences) and the single count of two very close/concurrent colonies (49 occurrences) are the most present. This represents error occurrences on a total of 3928 colonies counted on 122 boxes. The probability of error is therefore low and these occurrences can be easily corrected thanks to the double check that the technician makes at the end of the incubation on the display of the apparatus to validate the result.

Conclusion
This study confirms the capabilities of the ScanStation range of incubators for the detection of microbial colonies on membranes. It should be noted that in the current state of the software solution embedded in the instrument, the detection is only effective if black filtration membranes are used with the “black membrane” parameter of the instrument. However, it will be important to test several references of black membranes in order to choose the one that allows the development of the largest part of the microflora to be detected. This unavoidable step remains, however, a fairly tolerable validation for a laboratory since it only requires the verification of a precise element and not the whole technique.
The detection/counting performances are moreover largely sufficient since the detection is quantitative for colony numbers greater than or equal to 150, which remains the maximum value defined on a membrane in microbiological analysis.
During these tests, various detection and counting errors were identified. These errors are still minor compared to the volume of plates analyzed. The continuous software improvement proposed by the supplier will probably enable a major part of them to be resolved. These errors are all the more to be put into perspective, as in addition to their low occurrence, at the end of incubation, a verification of the photographic analyses and colony detections are checked by a technician who will be able to detect and correct these counting anomalies through a computer interface where the slightest modification of the analysis is traced and secured.
It is therefore a real technical solution in line with (1) the requirements of data integrity, (2) the need for accuracy of the analysis result, (3) which will free up considerable technical time that can be dedicated to ever-increasing activities in microbiological analysis laboratories on a release basis.
Share article
Sébastien Bonot – Aptar Pharma
Gautier Varin – Aptar Pharma
Manon Laborie – Interscience
Glossary
FDA: Food & Drug Administration
SDA: Sabouraud Dextrose Agar
TNTC: Too numerous to count
TSA: Tryptic Soy Agar
CFU: Colony forming unit
References
Corry J. E. L., Jarvis B., Passmore S., Hedges A., 2007, A critical review of measurement uncertainty in the enumeration of food micro-organisms, Food Microbiology, 24, 230-253.
FDA, 2018, Data Integrity and Compliance With Drug CGMP Questions and Answers Guidance for Industry, https://www.fda.gov/regulatory-information/search-fda-guidance-documents/data-integrity-and-compliance-drug-cgmp-questions-and-answers-guidance-industry
Fowler, J.L., Clark, W.S., Foster, J.F., Hopkins, A., 1978. Analyst variation in doing the standard plate count as described in Standard Methods for the Examination of Dairy Products. J. Food Prot. 41, 4–7.
Peeler, J.T., Leslie, J.E., Danielson, J.W., Messer, J.W., 1982. Replicate counting errors by analysts and bacterial colony counters. J. Food Prot. 45, 238–240.