Sommaire
- Vaccines & Endotoxin, a Challenging World.
- Méthode alternative d’identification. Exemple d’une implémentation d’un système Maldi Tof au laboratoire de contrôle.
- Implantation d’une méthode microbiologique alternative et polyvalente au laboratoire. Exemple du BACTALERT 3D®
- From Variable Operator Numeration To The Standardized 3P® Station Automated Colony Counting On Environmental Monitoring Culture Media Plate
- Analyse de la charge microbienne en temps réel pour prévenir les risques de contamination et mieux contrôler les procédés.
- ISO 22519 PW/WFI production systems standard: Quality aspects.
- Eau pour préparations injectables en vrac (productions, coûts). Étude de cas axée sur la qualité microbiologique.
- Coming to basics on rouging.
- Winning combination: Reducing particles in RTU packaging by aligning lid material, adhesive and sealing parameters in a holistic approach.
From Variable Operator Numeration To The Standardized 3P® Station Automated Colony Counting On Environmental Monitoring Culture Media Plates.
Regulatory requirements for cleanrooms classification and monitoring are detailed by EU GMP (Annex 1) or the FDA cGMP Guidance for industry, USP<1116>(1), ISO 14644 & 14698 and PDA – TR N°13. Those reference documents describe the Environmental Monitoring (EM) as a key element to ensure that an aseptic processing area is preserved in an adequate level of control while maintaining the full data integrity of the sample during the whole workflow of the analysis. The only technique to understand and follow the contamination of these critical clean rooms is the use of specific irradiated culture media plates that can recover the environmental flora.

The common accepted inspection practice of plates consists, after appropriate incubation temperatures and time, to enumerate colony forming units (CFU) by a qualified operator. The microorganisms should grow into distinct macroscopic colonies visible by a person without the support of magnifying devices.
But what is the limit that qualify a macroscopic object? And at which size can we consider the human detection accurate and reproducible? Would the use of automated microbiology methods for EM allow for better standardization and reproducibility of those numeration results? Also, how can we limit human errors and the risks of deviation and data integrity breach?
To answer those questions, we will present a global EM study in three chapters. At first, we will present the “BEADS STUDY“, a standardized methodology to evaluate the highly variable human performances of manual reading. In a second step, from a collaborative study between 4 pharmaceutical industries, the performance evaluation of a prototype automated system for incubation and numeration of colonies on plates at Sanofi Pasteur will be presented: the “INDUSTRIAL COLLABORATIVE STUDY“. And finally, the latest counting performance results of a fully integrated 3P® STATION compendial 4 Microbiologie 28 I La Vague N° 66 I Juillet 2020 incubation and numeration solution are presented in regard to the traditional reading: the “3P STATION PERFORMANCE STUDY“.
incubation and numeration solution are presented in regard to the traditional reading: the “3P STATION PERFORMANCE STUDY”.
1. Introduction
EM trend analysis rely on the accuracy of the results recovered on all Petri dishes used in the critical production environments. The analysis is quantitative, the results are expressed in numbers of CFUs with different specifications depending on the clean room classification, having the most drastic requirement of 0 CFU contamination in class A environment. The visual examination of these solid culture media is done by qualified operators who must comply with the microbiological best laboratory practices described in the USP chapter <1117>(2). In addition to a solid educational background, an internal training with initial and periodic evaluations must be followed to ensure accurate and reproducible results. Theoretically, there is no reason to have discrepancies on results between different trained operators. However, standardized results are difficult to achieve due to several factors such as the natural human variation in sight and evaluation or the huge variety of microorganisms colony morphology and growth patterns (sizes, shapes, translucent, proximal, spreading, …). The correct detection and enumeration of microbial colonies on plates is therefore in some cases a real challenge: this is illustrated by numerous FDA 483 forms (12) issued between 2011 and 2018 due to bad practices during Petri dishes inspection(3). Half of them are attributable to counting errors! This questions of the accuracy of the results generated during visual inspection Petri dishes.
• For this reason, we firstly developed the “BEADS STUDY” with a specific methodology to standardize and measure the quality of human detection on culture media dedicated to EM, including challenging events.
• Following the findings of this study, we developed an automated prototype, capable of incubating culture media plates and counting colonies (based on kinetic image analysis), and with the collaboration of several pharmaceutical industries during the “INDUSTRIAL COLLABORATIVE STUDY“, we defined the potential improvements to bring to this prototype to develop a final version.
• And finally, the final high capacity 3P STATION system was upgraded and qualified through the “3P STATION PERFORMANCE STUDY“, and integrated as part of the complete workflow of Environmental Monitoring through the 3P ENTERPRISE solution.
2. Materials and Methods
2.1 BEADS STUDY
Calibrated beads (from Cospheric LLC.) were used to mimic CFU on culture media plates (Table 1):
Petri dishes and culture media
A specific medium with no growth performance was developed in an EM dedicated LockSure® Petri dish: its formulation was established to prevent microbial growth and cross contaminations (mix of antibiotics) while allowing multiple readings by several people with the same quality. The Petri dishes were poured with a volume of 30 mL and the final aspect of the plates was identical to a regular EM culture media.
Samples preparation
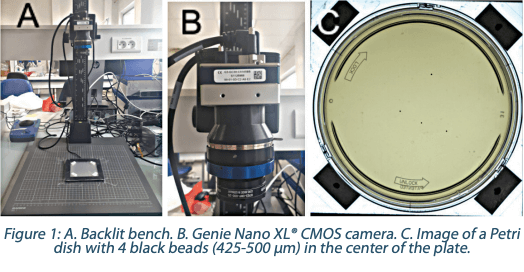
The different beads were directly dropped onto the agar plates. The bead size of 425-450 µm allowed the handling directly with tweezers. For the other sizes, beads were, beforehand, introduced in a sterile saline solution and diluted to achieve the expected concentration of beads in 50 µl of solution. The application on the agar was realized with the help of a CMOS image sensor camera (Genie Nano XL® C5100 Color, ref. G3-GC30-C5105 – Teledyne Dasla) positioned on a dedicated backlit bench (Figure 1).
Methodology A design of experiments (DOE) was realized to mix the following parameters (Table 2):
In total 1,088 plates were prepared, 79% negative and 21% inoculated with beads, with the purpose to mimic the higher ratio of negative plates observed in high grade clean room areas.
12 trained and qualified operators from 3 laboratories read independently each plate: 2 labs from bioMérieux (microbiology R&D team and culture media Quality Control team) and 1 pharmaceutical contract laboratory. The numeration was spread over a 5 days period to limit a potential impact of tiredness on the quality of the results. The plates examination was realized in the validated routine conditions with the possibility to use black and white backgrounds as well as additional lighting. No magnifying instrument were allowed.
A total of 13,056 results (10,296 on negative plates and 2,760 on positive plates) was generated. For positive results the total amount of data per parameter is explained below in Figure 2:
The deposit of accurate numbers of 53-63 µm white beads was impossible even using the high magnifiying camera, therefore those specific size and constrast beads were not analysed here in this study. Analysis were performed using the SAS Entreprise Guide v7.1 and Rstudio V1.1.442 softwares.
2.2 INDUSTRIAL COLLABORATIVE STUDY
This collaborative study was designed with 4 pharmaceutical industries to evaluate the performances of an initial 3P STATION Prototype with an incubation capacity of 100 culture media plates. The data shown in this study refers only to the Sanofi Pasteur evaluation.
Prototype equipment
The prototype equipment was a specifically customized ScanStation 100 from Interscience with preliminary algorithm version 1.17.
Culture media plates
COUNT-TACT 3P® Plates (CT3P, bioMérieux ref. 43699) and 90 mm TSA 3P® Plates (TSA3P, bioMérieux ref. 43169) were used in this collaborative study.
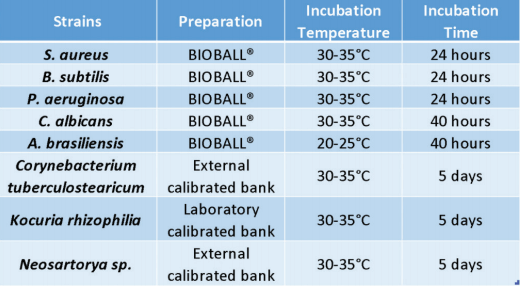
A DOE was set up with 5 pharmacopoeia strains and 3 environmental isolates (Table 3) prepared at 3 target concentrations (5, 25 and 50 CFU per plate) and then plated with five replicates split evenly between both CT3P and TSA3P Plates. This represents a total of 240 spiked plates and 4,697 individual colonies.
Each set of strains and plates was incubated and the remaining free positions in the incubator were filled by negative control plates. All plates were numerated by both the 3P STATION Prototype and the operators in order to measure and compare their reading performances.
Counting methodology
For each plate, numeration was done both by the 3P STATION Prototype with a kinetic approach (numeration every 1 hour during the incubation), and by 3 operators independently inspecting each plate with and end-point approach (after the full incubation cycle).
A “Reference Count” tool was developed to measure the performances of the operators and of the 3P STATION Prototype to improve as much as possible the detection algorithm.
The protocol to define the Reference Count for each plate after incubation is the following:
• The 3P STATION Prototype gives its final numeration and 3 operators independently enumerate the same plate,
• All counting results are reviewed and compared by the 3 operators,
• For each individual count, with the physical plate in hand and thanks to the video and images availability to “scroll back in time” the operators can better visualize colonies growing and agree afterward on a consensus to define the Reference Count for each plate.
Finally, each plate numeration result (operator or prototype solution) is compared against the Reference Count and assigned the false positives and the false negatives rates at the colony level where necessary.
Growth equivalency with automated incubation
To demonstrate the capability of the system to grow and recover microbial strains compared to a traditional incubation chamber, the 8 strains were spiked at a target concentration of 50 CFU/plate with five replicates on 90 mm TSA3P plates and incubated for 1, 2 or 5 days at 30- 35°C in parallel between the 3P STATION Prototype and the traditional laboratory incubator (warm room chambers). The numeration was done by one operator and the 3P STATION.
Cross-contamination evaluation
To show the absence of cross contamination in the 3P STATION Prototype incubator, 20 TSA3P plates spiked with high level 50 CFU/ plate of Aspergillus brasiliensis, were incubated together with 80 negative plates for 4 days at 20°C-25°C and 3 days at 30°C-35°C. Two air sampler tests were done with AIR IDEAL 3P inside the 3P STATION at the beginning and the end of the incubation, to check that no spores of A. brasiliensis contaminated the incubator during the incubation cycle.
2.3 3P STATION PERFORMANCE STUDY
3P STATION final design and performance metrics
Based on the initial findings of studies 1 and 2, a final version of the 3P STATION with a higher capacity of 300 culture media plates was designed (Figure 3).
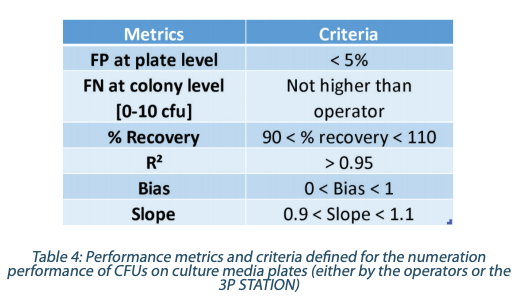
Performance metrics and criteria were defined for the 3P STATION instrument alone. For the False Negative rates determination (Table 4) the 3P STATION and one operator numeration were compared to the Reference Count for all the plates with less than 10 cfu/plate, which represents a total of 116 colonies.
Definition of the Metrics
False Positive (FP) at plate level occurs when the plate is negative and a numeration is given by the 3P STATION False Negative (FN) at colony level occurs when a colony present on the plate (Reference Count) is missed Percent Recovery is the ratio of the colonies detected on a plate by the 3P STATION over the Reference Count R² is the coefficient of correlation between the determined count and the Reference Count Slope & Bias are obtained with the regression analysis parameters between 3P STATION & the Reference Count as in the formula hereuner Y = aX+b with where Y = 3P STATION counts, X = Reference Count, and a = slope, b = bias
Methodology
A specific DOE was established to define the level of performances of the 300 culture media plates capacity 3P STATION.
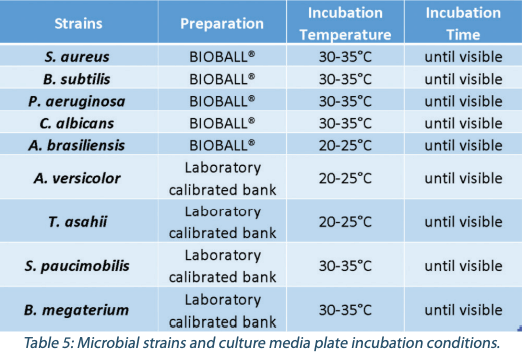
The verification was done on 88 culture media plates: one batch of 76 plates (86%) being either negative or inoculated with the same pharmaocopoeia strains as the Industrial Collaborative Study, and one batch of 12 plates (14%) being either inoculated with molds or with transluscent strains (Table 5). Colonies were inoculated between 0 and 50 CFU and evenly spread on each plate.
Numeration was done both by the 3P STATION and the operators, and compared to the Reference Count to assign the false positives and false negatives rates for each plate where necessary.
3. Results and discussion
3.1 BEADS STUDY
Analysis at the plate level
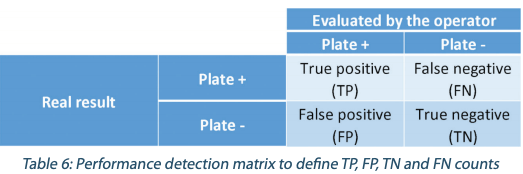
The objective of this analysis is to evaluate the natural perception of the operators during visual inspection and their capabilities to discriminate positive results from negative results. In this part, only the performance of detection of positive plates (plates with bead(s)) versus negative plates was evaluated, without taking into account of the accuracy of the enumeration: presence/absence triage test. The results are expressed as shown in the Table 6.
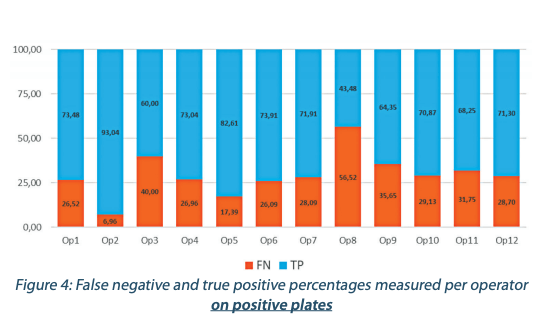
The performance of detection per operator is presented in the Figures 4 and 5. The Figures 4 and 5 show a huge variability in the results between the different operators. This tends to demonstrate that there is a huge level of subjectivity regarding the way a plate is considered positive or negative by an operator.
A possible explanation of these results could be the difficulty for the human eye to differentiate tiny objects in addition of the subjectivity to assess them as relevant or not. All the plated beads were equal or below a 500 µm size that can be considered as very small objects compared to microbiological colonies that can measure several millimeters, but also representative of slow growing stressed environmental isolates found in cleanrooms.
Therefore based on this beads study, it seems reasonable to extrapolate that the current reading and detection performances of trained operators of colonies on culture media plates is highly variable, generating a huge risk of false negative or positive results and a lack of data integrity due to the subjectivity of the test by design.
Analysis of the different parameters impact
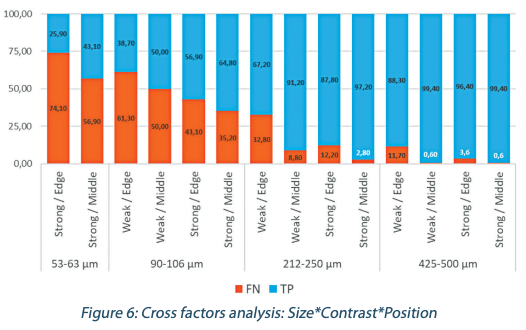
This part of the analysis is focusing only on positives plates to evaluate the accuracy of the enumeration at the bead level provided by the different operators. The results of the 12 operators were pooled together to evaluate the parameters that globally impact the visual inspection (Figure 6).
From the 3 criteria analyzed influencing the detection accuracy, the most impactful appears to be the beads sizes, and the less impactful appears to be their contrast.
• The figure 5 shows a clear correlation between the beads size and the detection performance. Below 212-250 µm up to 74% of incorrect results are measured, and the smaller the beads, the worse the numeration error level is.
• As for the location on the plate, between the middle or the edge position of the 212-250 µm beads, we observe a huge diminution of the percentage of correct results, respectively from 91.20% to 67.20%.
• The contrast influence is less significant but is well noticeable for beads around 212-250 µm with a logical better detection with more contrast.
Based on those results, we can reasonably consider that the accurate limit of detection for the human eye is close to 250 µm. Under this threshold, the percentage of missed objects becomes really high (more than 1/3 of false negative results). Therefore like the human operators, an automated detection technology should also be able to accurately detect 250 µm microbial with low variability.
3.2 INDUSTRIAL COLLABORATIVE STUDY
The objective of this study was to evaluate the performance of the 3P STATION Prototype in an industrial environment and to evaluate its performances to further identify potential improvements for the High Capacity 3P STATION. Out of the 4 collaborative sites, the data shown here are the ones generated by Sanofi Pasteur, at Marcy L’Etoile (France).
Counting performances on pharmacopoeia strains
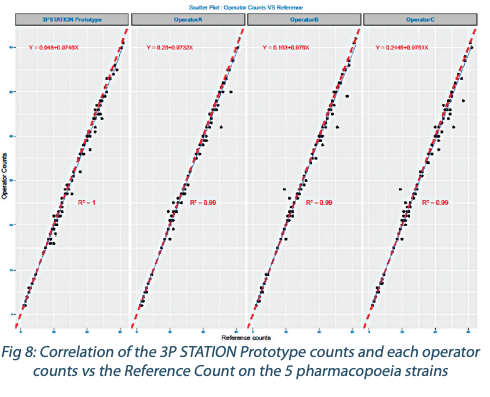
The regression analysis on each individual strain counted by the 3P STATION Prototype vs the operators (data not shown here) gives at least a R²>0.97. When analyzing all the strains together (Figure 7), the 3P STATION Prototype manages to obtain a R²=0.99 compared to the operators.
So the correlation between the 3P STATION Prototype and the operators is extremely strong and tight. And the dispersion of the count is even lower at low inoculum levels (<10CFU) specific to Class A cleanrooms.
And when performing the regression analysis on another angle on all the strain numerations vs the Reference Count, the correlation of the 3P STATION Prototype with an R²=1 was superior to that of the operators who have a tendency to under-numerate (Figure 8) with a higher dispersion.
The end-point reading of the operators seems to generate more counting errors than the kinetic method of the 3P STATION, and based on the R², the slope and the intercept, the 3P STATIONS brings more robustness and reliability of the results.
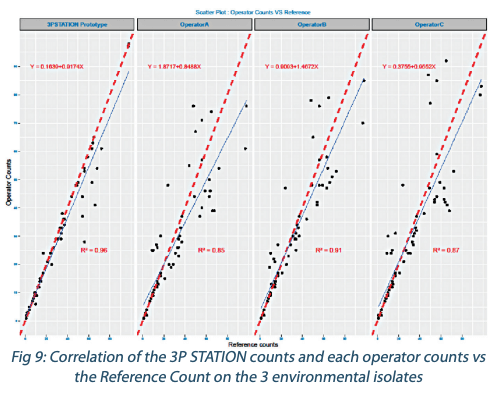
Counting performances on environmental strains
When performing the regression analysis on the environmental strain numerations by the operators and the 3P STATION vs the Reference Count, the correlation of the 3P STATION with a R²=0,96 is again far superior to that of the operators who have a tendency to undernumerate (Figure 9) with a much higher dispersion on higher counts. This confirms the initial findings that end-point reading by the operators seems to generate more counting errors than the more accurate kinetic method of the 3P STATION.
Growth equivalency with automated incubation
The average recovery rate between the 3P STATION and the operator numeration after incubating the plates in the traditional incubator was 107%, in accordance with the performance criteria set at 70% recovery rate acceptance.
Therefore, it is accepted that the incubators capacities to recover strains are statistically the same between the 3P STATION and Sanofi Pasteur’s current incubation chamber.
Cross-contamination evaluation
After the 7 days of incubation of the 20 spiked culture media plates in the 3P STATION, no contamination by Aspergillus brasiliensis was found on any of the 80 negative plates. One plate was found to be contaminated, but with a different strain that was identified as Micrococcus luteus sensu lato and investigated to be an operator contamination. And the active air sampling after the incubation cycle didn’t reveal any Aspergillus brasiliensis contamination either. Therefore the absence of cross-contamination in the incubator was demonstrated.
Note : in case it would be required, 2 specific protocols exist for either the cleaning or the decontamination of the incubator.
3.3 3P STATION PERFORMANCE STUDY
The objective of this study was to determine the performance of the High Capacity 3P STATION after the optimization brought to the solution thanks to the collaborative study.
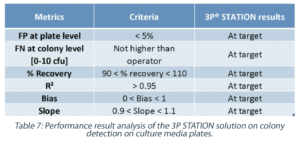
The analysis of all the defined metrics was therefore performed on the 88 plates (Table 7), and the False Negative rate was compared between the 3P STATION and the operators.
False Positive (FP) rate
The FP rate measured at the plate level – triage between negative and positive plates – was much lower than the acceptable specification, and well within the acceptance criteria set for the system.
False Negative (FN) rate
When comparing the operators and the 3P STATION performances, it is clear that the automation of the reading and the reproducibility of the algorithm allows for better performances with the 3P STATION. It is especially interesting to note that for low contamination rates (<10 CFU/plate) as it could be found in Class A & B cleanrooms, the operators had a FN rate above 5% when compared to the Reference Count, while the 3P STATION showed a much lower FN rate.
The conformity of the Recovery, R², Bias and Slope criteria demonstrate a highly reliable, robust and perfectly aligned 3P STATION solution compared to the Reference Count. All the criteria were at target with the defined specifications, and with a FN rate much lower than the operators, the 3P STATION is a well-adapted automated compendial method for EM plate numeration, as good or better than the human detection capability.
Conclusion
In 2011, Sutton published an article about accuracy of plate counts showing the high variability of the operators and that microbiology itself is a real limit to generate reliable data(4). In this paper, the correlation between the estimated error rate and the observed low plate count is clearly demonstrated. With the data of our BEADS STUDY, we demonstrate the natural variability of skilled operators that are performing the visual inspection of Petri dishes dedicated to EM. In the case of challenging detections, there is a part of subjectivity in the discrimination of small objects. We can then understand the impact of the background noise level on the quality of the detection. Depending on how operators identify objects as relevant or not, the balance of false positive and false negative can be dramatically different. On top of the natural differences of people performing the EM plates inspection, the study shows common influencing factors, the main one being the beads size. Reasonably, we can consider that the accurate limit of detection for the human eye is close to 250 µm. In addition to the size, the position and the contrast may play a role in the quality of the detection. EM visual inspection is a manual task, performed by people with variable performances by nature, creating issues of False Positive of False Negative numerations of colonies on plates. Improving the data quality, reliability and integrity from EM sample results can only be achieved with the implementation of automated compendial methods. As an opportunity of change and improvement, the Pharmaceutical industry should already consider automated solutions, replacing the traditional plate enumeration by automated incubators and plates counters. We have shown in our INDUSTRIAL COLLABORATIVE STUDY and our 3P STATION PERFORMANCE STUDY that the current EM culture media plate inspection method with operators (and sometime with a double reading) is not the state of the art anymore, and that automation should be the way to go forward. Newly available automated technologies allow to reduce drastically the level of False Positive and False Negative results from EM counts, while still maintaining the highest level of data integrity and compliance thanks to the digitalization of the information.
Aknowledgement
We warmly thank Laurent Leblanc, Lisa Mallam and Katia Imhof for their key contribution to this article. We also thank Clément Dilas, Matthieu Ribon, Laura Bailac and Franck Guilloteau who contributed to this project and helped in the design and roll-out of the studies and the analysis of the data.
Partager l’article

Arnaud PARIS – BIOMERIEUX
arnaud.paris@biomerieux.com

Lucile PLOURDE-OWOBI – SANOFI
lucile.plourde@sanofi.com
Références
1. GUSP, “1116 Microbiological Evaluation of Clean Rooms and Other Controlled Environments”, USP 34, United States Pharmacopoeia.
2. USP, “1117 Microbiological Best Laboratory Practices”, USP 34, United States Pharmacopoeia.
3. FDA U.S. Food & Drug Administration; Waring letters References: 320-11-013, 320-11-015, 320-12-019, 320-13-09, 320-13-09, 320-16-08, 320-17-01, 31-17, 320-18-35, 320-18-55; 483 Forms References: 3005531475, 3008386908
4. Scott Sutton, “Accuracy of Plate Counts”, Journal of Validation Technology, Vol. 17 n°3, pp. 42-46, 2011
Glossaire
GMP: Good Manufacturing Practices
EM: Environmental Monitoring
CFU: Colony Forming
Unit 3P: Pharmaceutical Proven Performances
DOE: Design Of Experiments